|
|
Taxonomy
| Click
on Systematics for a printable version of this article. |
Systematics:
Naming Organisms and Reconstructing the
Evolutionary History of Life
|
In
the last paragraph of The
Origin of Species Charles Darwin (1809-1882)
reflects eloquently on the common ancestry of life
on Earth. (1809-1882)
reflects eloquently on the common ancestry of life
on Earth.
“There
is a grandeur in this view of life, with its several powers,
having been originally breathed by the Creator
into a few forms or into one; and that whilst this planet
has gone cycling on according to the fixed law of gravity,
from
so simple a beginning endless forms most beautiful and most
wonderful have been, and are being created (Darwin, 1859/1993,
pp 648-649).” Click on picture to the right.
Continuity of Life
Charles Darwin saw a continuity
to life through his theory of evolution by natural selection. This same
continuity was echoed in
the work
of the German physician Rudolf Virchow (1821-1902). In 1852
Virchow hypothesized that cell division could account for cell
reproduction. In his 1855 classic work Die Cellularpathologie he
writes, “Omnis cellula e cellula” (all
cells arise from cells). The Cell Doctrine is a cornerstone
of modern
Biology and in the light of evolution indicates common ancestry
through cell division.
In adult humans each
of the more than ten trillion cells can be traced back to
the original unicellular
zygote. The zygote itself was the product of a sperm fertilizing
an
egg cell. The sperm and egg cell can be traced back to the
zygotes from which they arose. This simple thought experiment
takes us back from one generation to the next. There exists
an unbroken continuity to life; we can trace all of our cells
back to the very first cells that existed on Earth. Thus,
we can infer from the Cell Doctrine that all organisms can
trace
their cells back to the very first cell from which all life
arose some 3.5 billion years ago (deDuve, 2002, pp 9-10).
Binomial Nomenclature
In science, taxonomy
is the formal classification of organisms. Today, taxonomic
systems strive to cluster organisms into natural
hierarchical groupings based upon morphology and phylogeny.
Organisms are classified using a system developed by the Swedish
scientist Carolous Linnaeus (1707-1778). Prior to Linnaeus
the names of organisms consisted of descriptive phrases.
These
phrases were called polynomials. Linnaeus was attempting to
classify life on Earth in his book Systema Naturae,
first published in 1735. Linnaeus used polynomials, but also
provided a condensed
two-word Latin name for each organism. This two-word system
is called binomial nomenclature and is still used today.
Binomial
nomenclature is used to give each organism a scientific name.
The first word is the Genus (plural genera), which is always
capitalized. The second word is the species and is always written
in lower case. The generic and specific names are either italicized
or underlined. The name for humans is Homo sapiens. Once
the scientific name of an organism is introduced in a document
it can be abbreviated thereon.
The abbreviation for Homo sapiens is
H. sapiens. The domestic dog is Canis familiaris
(abbreviated C. familiaris).
In 1942 the famous evolutionary
biologist,
Ernst Mayr (1904-2005), proposed the biological species concept.
A biological species is a group of actually or potentially
interbreeding natural populations that are reproductively
isolated from other such groups. Identifying a species can
be problematic. Breeding
habits are not always known and less than 20% of living species
are properly described.
Many living species are described using morphology. Fossil
species are defined using morphology and phylogeny.
Linnaeus
grouped taxa (singular taxon) into hierarchical groups. From
most specific to most general we have: species, genus, family,
order, class, phylum, and kingdom. It was clear to Darwin
that the pattern of hierarchical groups in the Linnaeon system
mirrored
the splitting pattern of evolution. After Darwin taxonomists
were trying to form natural groups reflecting evolutionary
history.
Classification is always
changing as new fossil and molecular data are gathered. The
desire to form groups
based
upon evolutionary relationships has led to the development
of several contrasting taxonomic schools over the years.
Evolutionary taxonomy, numerical taxonomy, and cladism
were all developed
in an attempt to uncover evolutionary relationships. All
of these systems go beyond traditional taxonomy in that
they strive
to understand the diversity of organisms and the relationships
among them. The science that studies biodiversity is called
systematics. Systematics combines traditional taxonomy, phylogeny,
and biogeography to understand the evolutionary history of
life on Earth. Of the three competing systems mentioned above
we will focus
on cladism as it has become the standard for working scientists.
For a discussion of all three systems see Bringing
Fossils to Life by Donald Prothero.
Cladistics & The Tree of Life
Multiple
lines of evidence, including morphological data, biochemistry,
and
DNA sequencing support that all organisms are genetically
related. A single branching tree of life or phylogeny connects
all living and extinct forms of life. Evolutionary relationships
between organisms are established through cladistic analysis
of morphological data and molecular phylogeny reconstruction.
The German Entomologist Willi Hennig (1913-1976) developed
cladistics or phylogenetic systematics in the 1950’s.
Cladistics became well known when Hennig’s 1950 Phylogenetic
Systematics, originally published in German, was translated
to English in 1966 (Prothero, 1998, p. 46). Cladistics is a
school of taxonomy that establishes evolutionary relationships
based upon shared derived characteristics or evolutionary novelties.
Using cladistic analysis, paleontologists will construct a
cladogram, which is a branching diagram that shows evolutionary
relationships
between organisms. Three definitions are important for understanding
how a cladogram is constructed.
Shared primitive characters
or traits (symplesiomorphies) are characteristics shared by
all members of the group being compared. Shared derived characters
or traits (synapomorphies) are characters shared by only some
members of the group. Shared derived traits represent evolutionary
novelties that are unique to the immediate lineage being considered.
Primitive and derived characters are defined by the problem being
solved.
For
example,
hair and
mammary glands would be derived characters at the class level
for mammals, but a primitive character at the level of mammalian
orders or families (Prothero, 1998, p.48). An outgroup is an
organism which is only distantly related to the others and
is placed on the cladogram as a comparison.
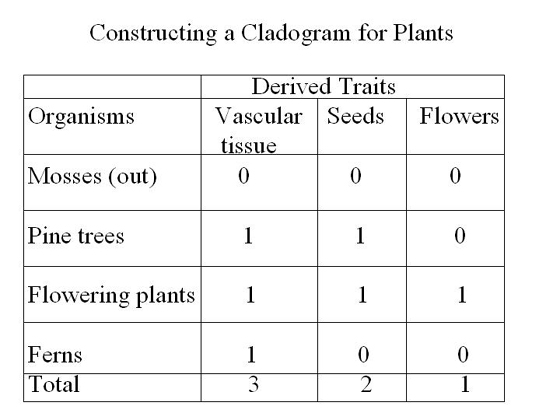
Constructing A Simple Cladogram
Let’s construct
a simple cladogram for plants to better understand cladistics
(Johnson & Raven, 2001, p. 328). In this table, the traits
for the out-group are marked with a zero. Traits not found
in the
outgroup
are
considered
derived
traits and are marked with a 1.
|
 |
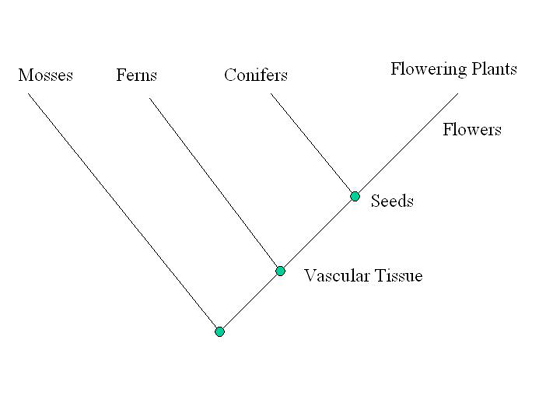
| Starting with a diagonal line, the out-group is placed on the
first branch. Just past the first branch, the most common derived
trait is listed; in this case vascular tissue is composed of
tube-like cells. The branching point or node on a cladogram marks
the point where shared derived characters arose. Next, the second
most common derived trait is determined, which in this case is
seeds. Ferns lack seeds and are thus placed on the second branch.
The third most common derived trait is flowers. Conifers do not
have flowers and are thus placed on the third branch. Flowering
plants are placed at the end. |
 |
Monophyletic
Groups
Cladograms show the closeness of relationships or how recently
two groups shared a common ancestor. The distances between
nodes are relative not absolute, so a cladogram is not an evolutionary
tree. The cladogram, supported by unique shared derived characters,
shows only that two taxa, branching from a common node, are
closely
related sister groups. A sister group consists of two lineages
sharing a common ancestor from which no other lineages have sprung.
Three
kinds of cladistic groupings are recognized. A
monophyletic group or
clade
arise from
a single
ancestor and include all the living and fossil descendants
of that ancestor (Benton, 2005, p. 32). Many familiar groups
are monophyletic such as
the phylum Chordata or subphylum Vertebrata. The members
of a clade or monophyletic group share at least one derived
character.
A goal of cladistic analysis is to identify monophyletic
groups because they are natural groups, true to phylogeny.
Many traditional
classifications include non-monophyletic groupings, although
they
are avoided whenever
possible.
A
paraphyletic group is a taxon that includes some but not
all of the descendants of a common ancestor (Prothero,
1998, p. 422). In a paraphyletic group some of the descendents
have lost the derived
trait.
The class Reptilia is a well-known paraphyletic group. Reptiles
most likely arose from a common ancestor, but the group
excludes birds (class Aves), which are descended from reptiles.
The term evolutionary grade is sometimes used to denote
a paraphyletic group. The class Reptilia has been replaced
by the class Sauropsida, which is monophyletic because
it includes birds.
A
polyphyletic group is a taxon that includes groups from
two unrelated
lineages (Prothero, 1998, p. 423). In a polyphyletic
group the apparent derived character
is actually the result of
convergent
evolution
and
the common
shared
ancestor
does not possess the feature. Grouping elephants, hippos,
and rhinos into pachyderms is an example of a polyphyletic
grouping (Benton,
2005, pp 31-32). Combining mammals and birds based upon
the characteristic of being warm-blooded represents a polyphyletic
grouping.
Cladograms are Testable
Cladistics
has become the major tool for scientists studying evolutionary
relationships and has had a profound affect on traditional
classification systems. Cladistics changed the focus
from finding ancestors
to analyzing patterns of shared specializations. The power
of a cladogram and the reason for its success is that
it is a scientific
hypothesis
that
can
be tested
by looking
at additional character states or additional taxa (especially
outgroups)(Prothero, 1998, p. 49).
Molecular Phylogenetics
Extinct and extant organisms in The Tree of Life
are connected to one another by their genomes. Thus,
molecules record evolution and can be used to establish the degree
of relationship between different organisms. Molecular phylogeny
consists of constructing patterns of evolutionary relationships
by comparing the
proteins and nucleic
acids of different organisms.
Multiple
methods are used to compare the distances between sequenced
nucleic
acids
(DNA and
RNA)
or proteins.
A molecular
tree of relationships is produced from the aligned gene
or protein sequences. The idea that molecules may mutate
at
a predictable
rate has led to the concept of the molecular clock. Thus
differences in molecular structure can be turned into
time of divergence
(Benton, 2005, pp 34-35). Molecular phylogeny reconstructions
represent
an independent approach
to
discovering phylogeny because they use genetic comparisons
instead of morphological differences as in cladistic
analyses.
In many cases, molecular phylogenetics has confirmed
evolutionary histories inferred by traditional phylogenetic
methods
(Lewin, 1997, p. 53). In other instances it has been
at odds with
morphological techniques. Molecular phylogenetics has
even solved problems
for which morphological techniques had no answer. Molecular
phylogenetics has influenced the classification and evolutionary
histories
of
many organisms. Molecular data broke up the Kingdom Monera,
provided evidence in support of the endosymbiontic theory
for the origin
of eukaryotic organelles, unraveled the origin of Australian
song birds, and helped to uncover the evolutionary
history of humans and other apes as well as change their
family taxa (Lewin, 1997, pp. 19-85).
Molecular
data support a growing body of evidence for the importance
of horizontal or lateral
gene transfer. Horizontal gene transfer or HGT is known to
be important
in the evolution of prokaryotes (Bacteria & Archaea). HGT
may also be a significant evolutionary factor for unicellular
eurkaryotes. So, the vertical gene transfer supposed by a tree
of life analogy is not the whole story. Some scientists argue
that an evolutionary web or net reticulating from the three
domains
of Bacteria,
Archaea,
and
Eukayota would
be a more useful analogy for the evolution of life than
a tree. Dr. W. Ford Doolittle points out that while some evolutionary
relationships are tree-like, many others are not (Lawton, 2009,
p. 39).
The Great Chain of Being & The Tree of Life
In the time of Linnaeus, scholars attempted to group organisms
from the simplest to the most complex in a hierarchy that reflected
the
fixed orderliness
of creation. In so doing, they were trying to discover anatomical
clues that linked life into a Great Chain of Being; revealing
the pattern of divine creation. Today we still look for similarities
and differences in form and structure (morphology) and use
some
of the same basic taxonomic methods. Since
the time of Linnaeus, much has been added to human knowledge
regarding relationships among life-forms that inhabit Earth.
We have greater awareness of the history of life on Earth as
it is revealed by the fossil record, extinction, mechanisms for
evolution, geologic changes in Earth's crust as well as the patterns
and molecules that guide inheritance.
In our time, scholars attempt to recover evolutionary history
using cladistic
analysis of morphological data and molecular phylogeny reconstruction. In
so doing, we are striving to recover The Tree or web of Life, a
phylogeny connecting all living and extinct forms of life on
Earth.
|
Bibliography
|
Benton,
M.J. (2005) Vertebrate Palaeontology [3rd Edition].
Blackwell Publishing: Main, USA.
Berra, T.M. (2009). Charles
Darwin: The Concise Story of an Extraordinary Man. Baltimore: The John Hopkins University Press.
Darwin, C. (1993). The
Origin of Species: By Means of Natural Selection or The Preservation
of Favored Races in The Struggle
for Life. New York: The Modern Library (Originally published
in1859).
De Duve, C. (2002). Life
Evolving: Molecules, Mind, and Meaning.
New York: Oxford University Press.
Johnson,
G.B. & Raven, P.H. (2001). Biology: Principles & Explorations.
New York: Holt, Rinehart & Winston.
Lawton,
G. (2009). Why Darwin was wrong about the tree of life.
New Scientist, January, 2009,
34-39.
Lewin, R. (1997). Patterns
in Evoluiton: The New Molecular View. New York: Scientific American Library.
Prothero,
D.R. (1998). Bringing Fossils to Life: An Introduction
to Paleobiology. New York: McGraw-Hill.
|
|
















